Coupling efficiency and Full-length oligo product:
The successful synthesis of long oligomers requires that the chain elongating coupling efficiency be as close to 100% as feasible. Even with a consistent 99% coupling efficiency across all the steps in the synthesis of a 100-mer (n = 100; requiring 99 coupling reactions), the amount of full length product in the resulting pool of oligonucleotides is roughly around 37%. This theoretical value for the same is determined using the following formula:
% of full length product in the product pool = (coupling efficiency)(n – 1)
The amount of the full-length product is a function of the length of the oligo and coupling efficiency. When the amount of the full-length product falls below a certain level, obtaining the mass spectra of the full-length product become challenging. Our new EXTREmers synthesis platform does typically facilitate >99.5% coupling efficiency, allowing for the synthesis of long oligos (upto 200-mers).
Synthesis of sequences with low sequence variation:
Our EXTREmer synthesis platform allows for the synthesis of DNA oligonucleotides even with very low sequence variation. The sequence,
CGG GGG CAG CGT GGA CGG GGG GCT GGG GAT GGG CGG GGT GGG GGC GGA GGG CGC GGG CGG CGG GCA GTG GGG GGC GGG CGG CGG GGG GGT GTA GAT CGG GGC GGG CGG G,
a 109-mer with 87% GC content, synthesized using our oligo synthesis platform is one example of such sequences. The ability to synthesize such oligos enables us to assemble genes or GeneStrands gene fragments with stretches of low sequence variation.
Error-rate, Cloning Efficiency, and Sequence Length
Genes are now more frequently assembled using chemically synthesized, overlapping oligonucleotides. The overlapping oligonucleotides are ligated, extended, or 'filled' to create larger constructs or even genomes. Owing to mistakes in chemical synthesis and subsequent enzyme-based assembly processes, the assembled genes unavoidably contain errors. The average rate of the errors attributed to chemical synthesis typically range from 1 to 10 errors per kilobase pair of assembled DNA. The following graph on the left displays the theoretical probability of finding an error-free gene of different lengths at a given error rate.
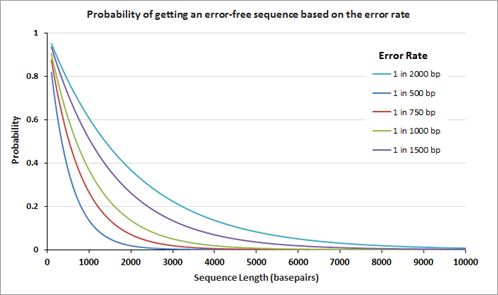
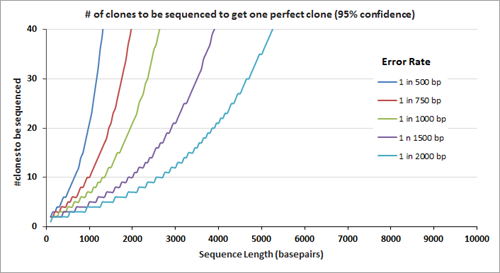
After gene assembly, cloning and subsequent sequencing of the isolated DNA from multiple clones allows a researcher to identify the clone(s) with the correct sequence. The number of clones that need to be sequenced to identify the one perfect clone also depends on the error rate; the graph on the right displays the number of clones that need to be sequenced (at 95% confidence level) before the 'one' is found. Clearly, the number of clones that need to be sequenced is fewer when the clones are assembled using the lower error rate oligos.
More recently, several enzyme-based error-removal methods have been developed to mitigate the propagation of synthesis errors incorporated during chip- and array-based synthesis. These error-removal methods essentially use enzymes to enrich the pool of the oligos with the desired oligos by selectively removing the 'undesired' oligos. The error-removal methods can be readily simplified when oligos with low error-rate are used in the initial assembly of the genes.
Extremely low sequence errors
EXTREmers, much like the other chemically synthesized oligonucleotides at our Oligo synthesis facility, are subjected to the conventional QC processes, i.e., mass spectrometry (and/or electrophoresis in case of EXTREmers), before being shipped to you. We are, however, aware that the MS- and Chromatography-based analytical techniques do not provide sufficient information about the sequence of the chemically synthesized oligonucleotides. The mass spectra of ACGGAGA is indistiguishable from that of ACGAGGA. Therefore, in a representative study, we used EXTREmer oligos in cloning and subsequent sequencing experiments to evaluate the sequence-errors present in the synthesized EXTREmer and the propagation of these errors during the gene assembly and cloning process.
| Length of Gene (bp) |
GC% |
Good Clones (%) |
Total kbp Sequenced |
Number of Seq. Errors |
Error Rate (%) |
| 617 |
65 |
45 of 64 (70%) |
39.488 |
21 |
0.05% |
| 1,020 |
41 |
10 of 20 (50%) |
20.400 |
11 |
0.05%
|
| 1,286 |
40 |
34 of 48 (71%) |
56.922 |
20 |
0.03% |
| 1,520 |
44 |
17 of 31 (55%) |
47.120 |
19 |
0.04% |
| 1,791 |
44 |
4 of 18 (22%) |
32.238 |
28 |
0.09% |
| 1,869 |
52 |
3 of 8 (38%) |
14.952 |
7 |
0.05% |
| 1,890 |
51 |
6 of 15 (40%) |
28.350 |
14 |
0.05% |
| 1,947 |
52 |
5 of 11 (45%) |
21.417 |
10 |
0.05% |
Genes of different sizes and GC contents were assembled using EXTREmer oligos. The sequences of the resulting genes were read either using the Sanger method or Next-Gen Sequencing methods. The graphs above display the Cloning Efficiency (A) and the error rates (B). Sequencing errors include insertions, deletions, and substitutions. No error-removal methods were employed during the assembly of these genes.
In summary, these results indicate that nearly 50% of the clones harbor a DNA of the correct size and 1 error is observed for a sequence lenght of 2,000 bp (the sequencing errors are further reduced when error-removal methods are used during gene assembly).
What do these results mean for your experiments? You will have to select fewer colonies during your cloning experiments and the likelihood of the sequence of the isolated DNA being the desired sequence is high! These features save you both time and money (used for the reagents and conducting the various molecular biology experiments).
Morever, our studies reveal that the number of sequence errors does not vary significantly based on the GC content of the assembled target sequence. The genes in the study below were assembled using PCR assembly with high-fidelity enzymes. Subsequently, error removal methods were used.
Effect of GC content on the errors (1500 bp gene)
| GC content (%) |
Error rate (%) |
Total kbp Sequenced |
| 30 |
0.0025 |
16.236 |
| 45 |
0.0038 |
13.284 |
| 60 |
0.0055 |
16.236 |
| 70 |
0.0041 |
14.760 |
| Average |
0.0040 |
60.516 |
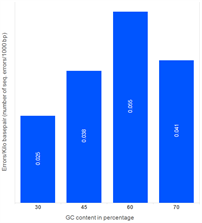
Sequence errors observed in target genes (length ~1,500 bp) containing different levels of GC content.
Please note that Genes and GeneStrands prepared at our Louisville gene synthesis labs are assembled using EXTREmers. Both these products are synthesized using low-yield, high-fidelity chemical synthesis protocols and therefore yield oligonucleotide products with very low-sequence errors.
Order Tubes
Order Plates