Mother Nature has optimized the sequence of genes through evolution. Further machineries that transcribe and translate the information contained in that gene sequence are optimized to generate the appriopriate levels of the enzymes in that organism. This optimization to generate the wild-type gene has occurred over several generations and ages. Why then should the DNA sequence of the gene be optimized.
Why Optimize?
When using a gene for protein expression, many times the wild-type gene is not optimized for maximum expression or the protein is expressed in a host that is not the original organism. By redesigning the gene, it may be possible to significantly increase the amount of protein expressed.
How is it possible to optimize a Gene sequence?
As most amino acids are encoded by more than one triplet, the number of possible DNA sequence versions for a single protein sequence is very high. Two amino acids are encoded by only one codon (Met and Trp), all other amino acids are encoded by more than one codon. E.g. Cys, Lys or Asn are encoded by two triplets, Ile is encoded by three triplets. Ala, Gly or Thr are encoded by four codons and there are three amino acids being encoded by six different triplets: Arg, Leu and Ser. On average an amino acid is encoded by 3 triplets, therefore a protein of 100 amino acids can be encoded by 3100 (= 5.2 x 1047) different DNA versions! Moreover, the translational machinery within different organism display different preferrences for the codons.
The Genetic Code
| First Letter |
Second Letter |
Third Letter |
| |
U |
C |
A |
G |
|
| U |
UUU |
Phe |
UCU |
Ser |
UAU |
Tyr |
UGU |
Cys |
U |
| UUC |
UCC |
UAC |
UGC |
C |
| UUA |
Leu |
UCA |
UAA |
Stop |
UGA |
Stop |
A |
| UUG |
UCG |
UAG |
Stop |
UGG |
Trp |
G |
| C |
CUU |
Leu |
CCU |
Pro |
CAU |
His |
CGU |
Arg |
U |
| CUC |
CCC |
CAC |
CGC |
C |
| CUA |
CCA |
CAA |
Gln |
CGA |
A |
| CUG |
CCG |
CAG |
CGG |
G |
| A |
AUU |
Ile |
ACU |
Thr |
AAU |
Asn |
AGU |
Ser |
U |
| AUC |
ACC |
AAC |
AGC |
C |
| AUA |
ACA |
AAA |
Lys |
AGA |
Arg |
A |
| AUG |
Met |
ACG |
AAG |
AGG |
G |
| G |
GUU |
Val |
GCU |
Ala |
GAU |
Asp |
GGU |
Gly |
U |
| GUC |
GCC |
GAC |
GGC |
C |
| GUA |
GCA |
GAA |
Glu |
GGA |
A |
| GUG |
GCG |
GAG |
GGG |
G |
Gene sequences, or segments thereof, having low variations in the arrangement or composition of the bases can lead to challenges in their synthesis and assembly; such sequences are classified by GENEiusTM as 'complex', because these features make the assembly of the custom Gene or GeneStrand gene fragments complex. Oligos containing long streches of a monomer, in particular stretches of the base G, have proven to be difficult to synthesize. When segments of the sequence repeat themselves, i.e., direct repeats, unintended binding and secondary structures can occurs. So too is the case when there are indirect repeats. i.e., a sequence and its reverse complement, in the sequence. GENEiusTM identifies such features in the entered sequences and provides recommendations that will allow you modify the sequence. To order a GeneStrand gene fragement or gene with all its 'complexities' intact, please contact our customer support at GenomicsSupport@eurofins.com. The turnaround time for the synthesis of complex genes or GeneStrands gene fragments is typically much longer than that for the standard genes or GeneStrands.
Features of sequences that make assmebly of Genes or GeneStrands complex include:
- Homopolymeric stretches of >18 base pairs
- Stretches of direct or inverted repeats
- Stretches of sequences with very high (>75%) or very low (<35%) GC content
- Segments of sequences that can result in critical secondary structures
4D™ Sequence Optimization with GENEius Software
GENEius is a powerful sequence optimization tool for protein expression from a gene. By optimizing in 4 dimensions, GeneOptimizer assures the best expression possible.
- Codon usage optimization
- Secondary structure avoidance
- Bad motif removal
- GC content optimization
Plus, Eurofins Genomics can add promoter/enhancer regions to your gene to improve protein expression.
1. Codon Usage Optimization
When expressing proteins in a host other than the original organism, the most-favored codons for specific amino acids can differ widely. Even if the host is the original organism, nature may not have created the gene in a way that fully expresses the protein.
GENEius will optimize the sequence of your original gene to use the most-favored codons possible for each amino acid.
2. Secondary Structure Avoidance
Strict codon optimization can result in unwanted secondary structure, greatly reducing the expression of your target protein. By adjusting the codon sequences used for your gene,
GENEius can reduce or eliminate secondary structure to assure maximum expression.
3. GC Content Optimization
Codon optimization can also result in an undesirable GC content.
GENEius gene sequences so that GC content is within the desirable range.
4. Bad Motif Removal
A sequence that is cleaved by your restriction enzyme will guarantee failed expression.
GENEius for these and other bad motifs and ensure that they are not found in your gene.
5. Promoters and Enhancers
GENEius software can also add promoter/enhancer regions to supercharge your gene for maximum protein expression.
Comparison of sequence optimization by GENEius with that of optimization algorithms In a comparative study, researchers at Eurofins Genomics compared GENEius with five major competitors for the optimization of jellyfish Aequorea victoria wild-type GFP for best expression results in E. coli. Resultant sequences were synthesized and subcloned into a modified pTrcHis (Invitrogen) for expression in E. coli TOP10 cells. Fluorescent signals of normalized E. coli cultures were measured with a fluorescence spectrometer, and the results are shown in the figure below.
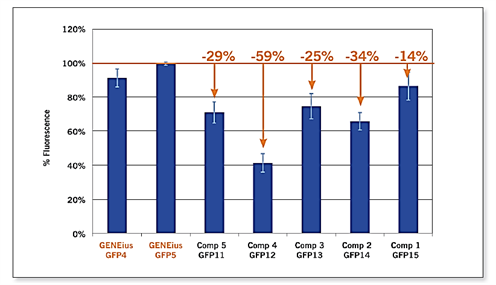
Figure 1: GFP fluorescence of optimized gene constructs. Constructs GFP11 – GFP15 were optimized independently by five competitors Comp 5 – Comp 1 respectively. Constructs GFP4 and GFP5 were optimized using GENEius.
As shown in the results, the optimization by GENEius produces significantly higher protein expression levels in E. coli than most of the popular codon usage adaptation and optimization software packages on the market. For a more in-depth look into this study, read the whitepaper on this page.
Considering the diverse application of GeneStrands gene fragments, it is not possible for us to discuss resolving all the likely complexities in a sequences. The following set of recommendations are suitable when using the GeneStrands gene fragments or Genes for cloning and expressing proteins. For other applications involving linear of circular dsDNA ensure that the modifications implemented for sequence optimization does not interfere with the intended application of GeneStrands gene fragments or Gene.
Homopolymeric stretch
A segment in the sequence of gene which encodes for an Arginine followed by 3 glycines can be the following: AGG GGG GGG GGG. However, contigous strands of G are notorious for creating problems in the synthesis of oligos. GENEiusTM will categorize this sequence as complex, with 'Hompolymeric Stretch' as the complexity. The recommendation for resolving such homopolymeric stretches is to limit the stretches of G's (or other repeating bases) to less than 8 bases in the segment.
| Amino acid |
X |
X |
X |
X |
X |
X |
Arg |
Gly |
Gly |
Gly |
X |
X |
X |
X |
X |
X |
| Initial Sequence |
NNN |
NNN |
NNN |
NNN |
NNN |
NNN |
AGG |
GGG |
GGG |
GGG |
NNN |
NNN |
NNN |
NNN |
NNN |
NNN |
| Resolved sequence |
NNN |
NNN |
NNN |
NNN |
NNN |
NNN |
AGG |
GGU |
GGG |
GGG |
NNN |
NNN |
NNN |
NNN |
NNN |
NNN |
Direct repeats
The DNA sequence of gene expressing a protein containing multiple poly-hisitidine tags (or other such repeating streches of amino acids) are likely candidates to have direct repeats. In the example sequence below, these repeats are at positions 4 and 28.
5'-NNNCATCATCACCATCACCACNNNCATCATCACCATCACCACNNN...3'
The two underlined fragments are direct repeats and can pose a challenge for the synthesis of the gene or GeneStrand. To resolve this you can change the code for one of the Histidines.
| Amino acid |
X |
His |
His |
His |
His |
His |
His |
X |
His |
His |
His |
His |
His |
His |
X |
| Initial Sequence |
NNN |
CAT |
CAT |
CAC |
CAT |
CAC |
CAC |
NNN |
CAT |
CAT |
CAC |
CAT |
CAC |
CAC |
NNN |
| Resolved sequence |
NNN |
CAT |
CAT |
CAT |
CAT |
CAC |
CAC |
NNN |
CAT |
CAT |
CAC |
CAT |
CAC |
CAC |
NNN |
Indirect Repeats
Segments of sequences can sometimes prove to be inverted repeats of each other. Such features can lead to the formation of unintended secondary structures. An 18-nt long arbitrary sequence starting at position 4 and its inverted repeat starting at postion 25 are included in this sequence: NNNCCAGTTATTACGAGATTTNNNAAATCTCGTAATAACTGGNNN.
| Amino acid |
X |
Pro |
Val |
Ile |
Thr |
Arg |
Phe |
X |
Lys |
Ser |
Arg |
Asn |
Asn |
Trp |
X |
| Initial Sequence |
NNN |
CCA |
GTT |
ATT |
ACG |
AGA |
TTT |
NNN |
AAA |
TCT |
CGT |
AAT |
AAC |
TGG |
NNN |
| Resolved sequence |
NNN |
CCA |
GTT |
ATT |
ACT |
AGA |
AGG |
NNN |
AAA |
TCT |
CGT |
AAT |
AAC |
TGG |
NNN |
GC content
Sequences with either very low or very high GC content can present challenges to the oligo synthesis and gene or GeneStrand assembly. These are typically indicated for the corresponding segment of the sequence. The number of G and C's within the window of the sequence must be reduced or increased based on the suggestion by GENEiusTM.