Direct sequencing of bacterial colonies using rolling circle amplification (RCA)
Direct Colony Sanger Sequencing (DCS) enables fast, reliable plasmid DNA sequencing directly from bacterial samples without the need for plasmid preparation. This approach accelerates the plasmid sequencing workflow and reduces hands-on sample prep. This webpage explores the uses, value, and applications of Direct Colony Sequencing. Eurofins Genomics accepts the following sample types for direct colony sequencing:
- Bacterial Colonies
- Glycerol Stock / Bacterial Culture
- Bacterial Pellet
Note: Only bacterial samples of biosafety level 1 are supported. Fungal colonies and other sample types are not accepted.
Sample Submission Guidelines
Please visit the Sample Submission Guide for information on how to prepare samples for direct colony sequencing.
About Direct Colony Sequencing and Rolling Circle Amplification
Rolling circle amplification (RCA) is a critical part of direct colony sequencing. It enables fast Sanger sequencing of plasmid clones without the need for plasmid preparation. The RCA method utilizes bacteriophage Phi29 DNA polymerase to exponentially amplify single- or double-stranded circular DNA templates. The isothermal amplification method produces microgram quantities of DNA from picogram amounts of starting material in a few hours.
An in vitro amplification of very small amounts of template DNA eliminates the need for overnight cell culture and conventional plasmid or M13 bacteriophage DNA purification. The proofreading activity of the Phi9 DNA polymerase ensures high fidelity DNA replication
The Direct Colony Sequencing service has been validated primarily for plasmids in E. coli colonies. In principle, the method also works with large constructs (e.g. BACs) and other organisms. Contact us for pilot testing and optimization. As a starting point, we recommend performing cell lysis or crude DNA extraction and adding 1 to 5 µL of the lysate/extract to the buffer in the 96-well plates.
| Highlights |
Benefits at a glance |
| |
|
|
Fast
Receive your sequencing results one day earlier for your plasmid clones as DNA preparation is not needed. Results are available the same day as samples are received
Flexible
It is easy to submit direct colony sequencing online. Users can select the sample type from any of the standard sequencing submission pages online.
Cost-Effective
Reduce your expenses on plasmid DNA isolation and let us sequence your bacterial colonies directly.
|
- Available for E.coli colonies or any other gram negative bacteria
- Up to 4 reactions per sample, per plate are possible
- High quality read lengths of up to 1,100 bases
- Results are available the same day as samples are received
- Free one time repetition in case of technical failure*
- Different sequencing result files can be selected per order
- A huge amount of standard primers are available free of charge
- Sequencing primers can be ordered or enclosed
- Easy re-order options from stored DNA samples
|
Value of Direct Colony Sequencing
Direct Colony Sequencing saves time. It sequences the plasmids/vectors directly from bacterial colonies without the need for plasmid preparation. You just need to pick the colonies and directly transfer them to one of the acceptable submission formats. The wells of the tube and/or plate contain buffer to preserve the colonies.
This fast and cost-effective solution can be used to quickly screen your DNA cloning experiments and select the candidate(s) for final confirmation via DNA preparation and sequencing.

Time and Cost Efficiency: DCS eliminates the need for DNA extraction and purification steps, reducing the time and cost associated with traditional methods. Researchers can directly sequence colonies, accelerating the genomic analysis process. This efficiency is particularly beneficial when dealing with large sample sizes or high-throughput applications.
Applications of Direct Colony Sequencing
Direct colony Sanger sequencing enables rapid characterization of plasmid-borne genetic elements from cultured bacterial isolates.
Typical applications include verification of engineered plasmid constructs, confirmation of known resistance or virulence genes carried on plasmids, and monitoring plasmid stability across laboratory, industrial, or environmental strains.
Process
You have prepared your DNA construct, cloned it into a vector and transformed E. coli with it. Now it is time to pick and grow the colonies and conduct minipreps in order to extract plasmids to send for sequencing. Confirmation of a construct’s presence and identification of the construct in the vectors is absolutely essential before you continue to work with the plasmids. However, this process is quite laborious and time-consuming, especially when there are a large number of colonies to check.
Rolling circle amplification process
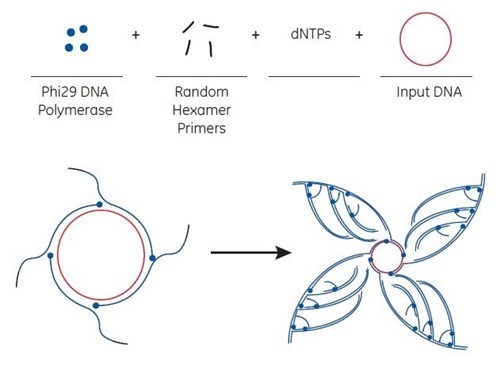
The basis for the Direct Colony Sequencing solution is the rolling circle amplification (RCA) technique that generates multiple copies of circular DNA. The starting material for amplification can be of various sources, such as from a small number of bacterial cells containing a plasmid, isolated plasmids and intact M13 phages to any circular DNA (including ligation reactions). Bacterial colonies can be picked from agar plates and added directly to the 96-well plate. Alternatively, microlitre quantities of a saturated bacterial culture or a glycerol stock can also serve as starting material.
RCA is performed at 30°C without thermal cycling. Depending on the source of starting material, amplification is completed in 6 to 18 hours. The amplification products are double-stranded concatemers of the circular template with high molecular weight. Concatemers are large DNA molecules that contain many copies of a specific DNA sequence.
For RCA, random hexamer primers anneal to the circular single-strand template DNA at multiple sites. The Phi29 DNA polymerase extends each of these primers. When Phi29 reaches a downstream extended primer, strand displacement synthesis occurs. Here, the original single-stranded DNA is replaced by the newly replicated strand. The displaced strand is then available to be primed by more hexamer primer. The process results in exponential amplification.
Note: when starting with M13 bacteriophage clones, the product is double-stranded DNA and can be sequenced with forward and reverse primers. DNA amplified by the method can be used directly without any purification.
The Direct Colony Sequencing service has been validated primarily for plasmids in E. coli colonies. In principle, the method also works with large constructs (e.g. BACs) and other organisms. Contact us for pilot testing and optimization. As a starting point, we recommend performing cell lysis or crude DNA extraction and adding 1 to 5 µL of the lysate/extract to the buffer in the 96-well plates.
References
Dean, F. et al., Genome Research 11, 1095–1099 (2001).
Lizardi, P. et al., Nat. Genet. 19, 225–232 (1998).
Estaban, J.A. et al., J. Biol. Chem. 268, 2719–2726 (1993).
FAQ
- How is a technical failure defined?
- A technical failure is defined as a chemical or technical (IT, machines) defect in our lab. In order to ensure that the sequencing quality is not affected by a technical failure, an internal quality is run with each plate (for that purpose well H12 should be kept empty, otherwise a quality control is not possible). In addition to that, a sophisticated software is analysing each plate considering parameters like quality values (QV) and reading lengths. If these parameters fall below a certain percentage across the plate an additional manual check is performed. If a technical failure is confirmed by this manual check, the plate is getting re-sequenced.
- What is the storage time?
- DNA and primer storage:
- Storage of template DNA for 4 weeks
- Storage of synthesised primers for 4 weeks free of charge
- Storage of synthesised primers for one year at additional cost
Data storage:
Secure online archive of all selected sequence data for 6 months.