INVIEW Transcriptome Explore
Reliable transcript identification and comparison between expression levels
INVIEW Transcriptome Explore is an attractive sample-to-result package that provides general characterization of a specific transcriptome using RNA-Seq. The product enables the comparative analysis of genes expressed at a given time or under specific conditions. The raw data is carefully analyzed by trained bioinformaticians to produce a clear and informative report of all sequencing results.
Applications
- Comparison studies of highly expressed genes
- Analysis of transcript levels from different samples
- Screening approaches where reference sequences are available
- Gene expression profiling in cancer
Highlights of INVIEW Transcriptome Explore
- Strand-specific cDNA libraries for sequencing with Illumina's technology
- 30 million single reads guaranteed
- Ideal for screening projects with high sample throughput
- Well suited to comparison studies of gene expression
Product specifications
- Processing of any number of samples, starting with one sample
- Preparation of random primed cDNA libraries
- RNA-Seq with leading sequencing technology
- Adaptable sequencing output in multiples of 30M reads
- High-quality BioIT analysis
- Final report with ready-to-publish data
- Available under ISO 17025 accreditation
- Requires a well-known reference genome/transcriptome
INVIEW Transcriptome Explore offers a powerful method for profiling and comparing transcripts in order to explore gene structure and function. The NGS-based approach offers general transcriptome analysis using RNA-Seq. The transcriptome-sequencing service extracts valuable information from carefully prepared random primed cDNA libraries. Following thorough BioIT analysis (Fig. 1), a comprehensive report of all results can be delivered in rapid turnaround times.
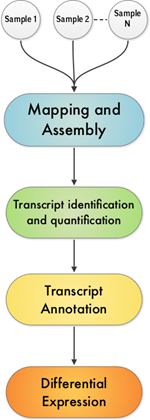
Fig. 1 BioIT analysis involved in the determination of gene expression levels
For projects without a reference genome, de novo transcriptome sequencing based on PacBio technology can be added to your project. See section below on "Related Products".
Learn more about the INVIEW Transcriptome Explore package
Starting Material
Accepted starting material for INVIEW Transcriptome Explore
- Total RNA from various sources
- rRNA-depleted total RNA
Accepted starting material for RNA isolation
- Tissue
- Cells
- Body fluids
- FFPE tissue
Please note that only S1-classified material is accepted for RNA isolation ordered online. More information about the current rules for classifying biological material can be found here. Please contact us for further information on isolating RNA from material classified as S2.
Specifications
Library types
- Standard eukaryotic library - mRNA is poly-(A) enriched and subsequently fragmented. cDNA synthesis is performed with random hexamer priming.
- Standard prokaryotic library - ribosomal RNA is removed by ribo depletion. Random hexamer primers are used for cDNA synthesis.
- Library with FFPE RNA - RNA isolation from Formalin-Fixed Paraffin Embedded (FFPE) samples. An optimised workflow including ribodepletion allows for reliable gene expression analysis and SNP detection.
Sequencing platform
Sequencing mode
- 50 bp single reads
- 30 million single reads guaranteed
Bioinformatics offered for transcriptome analysis
- Semi-automatic mapping against one reference
- Identification of potential exon-exon splice junctions
- Identification and quantification of transcripts
- Merging of identified pieces to full-length transcripts
- Transcript annotation based on known annotations
- Determination of expression levels
- Pairwise comparison of expression levels and determination of significant fold differences
- Detection and annotation of SNPs and InDels
- Annotation of detected SNPs and InDels that are registered in dbSNP
- Allocation of protein-level effects that are registered in Ensemble
Deliverables of bioinformatic analysis
- Alignment file (BAM)
- Gene expression table including FPKM value (tsv)
- Combined gene expression table of all samples (tsv)
- Table of top genes expressed (tsv)
- Table of pairwise differential gene expression including foldchange and p-value (tsv)
- Table of genes expressed differently between pairs (tsv)
- SNP and InDel tables including annotated variants and effects (vcf, tsv, bed)
- Comprehensive Data Analysis Report (.pdf)
Delivery time
- 15 days for up to 20 samples; an additional 6 days for every 16 additional samples; 96 samples upon request