GENEius - the intelligent sequence optimization software with enhanced functions.
Our proprietary software GENEius enables us to design the sequences of products such as GeneStrands and Genes so that they enable the best protein expression in different organisms.
As a result of redundancy in the genetic code, a single protein can be encoded by several distinct DNA sequences. Therefore, to design a gene to express protein in a heterologous organism, we have to choose from an enormous number of possible DNA sequences. A 100-mer protein sequence acids can be encoded in any one of the 3100 or 5.2 x 1047 distinct DNA sequences (assuming each amino acid is encoded on an average by three codons). The algorithm embodied in the GENEius software optimizes the input sequence against various parameters and generate sequences that offer improved protein expression in the host. During sequence optimization, GENEius calculates a score based on the following iterative steps:
- Uses data from the Kazusa Codon Usage Database (or that provided by the researcher) to increase the codon adaptation index of the seqeunce.
- The codons in the sequence are then harmonized, i.e., codon usage in the sequence is assigned based on the frequency of their distribution in highly expressed genes, while the very rare codons are completely avoided.
- 'Bad Motifs' like artificial splice sites, unspecified transcription factor binding sites, unwanted restriction sites, etc., are removed.
- Direct and inverted repeats in the sequence are removed to reduce the number of possible secondary structures—these not only make the synthesis of the oligo difficult, but also reduce efficiency of transcription and translation in several expression hosts.
- The GC content is distributed equally across the segments of the sequence to avoid GC-rich segments in the sequence.
Sequences that do not make the cutoff score are dropped. The resulting sequences are devoid of the 'bad motifs' and are optimized for protein expression in the target host. With GENEius, the efficiency of gene synthesis and further downstream applications are remarkably improved!
To use or evaluate the optimizing power of GENEius on your sequence of interest for free, you can use the 'GENEius Light' version that you can access here. Please note that this link opens a new window and takes you to a new site. You will have to create a separate account here.
The GENEius software is designed and developed for Eurofins Genomics by BioLink Informationstechnologie GmbH.
Complex segments in the sequence make synthesis of oligo challenging and reduce the efficiency of transcription and translation. Complexities in a sequence include:
- Streches of direct or inverted repeats (>20 bp)
- homopolymeric streches (>18 bp)
- Streches of sequence that can result in critical secondary structures
- Streches of sequence with very high (>75%) or low (<35%) GC content.
Secondary structures reduce the synthetic yields of DNA oligos while also reducing the translational efficiency, since these secondary structures are then adopted by the mRNA. Any Genes or GeneStrands that contain these features are considered 'complex' and take longer to assemble.
These complexities can also be removed when GENEius is used to optimize the sequence. GENEius, which is integrated into the order wizard used for ordering Genes or GeneStrands, allows for seamlessly removing the complex features in a sequence. The use of GENEius is optional, and your sequence of choice can be assembled—please contact a sales representation for your region for inquiring about feasibility and potential turnaround times.
What are the benefits of using GENEius for optimizing the sequence of Genes or GeneStrands?
GENEius—the beauty studio for genes.
No single Gene sequence is suitable for every project. With GENEius, you can tailor the sequence of your genes for optimal performace in your specific requirement.
Natural genes have evolved over generations to maintain a balanced expression for all celullar genes. As a result, natural genes often contain bad motifs; like hairpins, that restrict the usability of the gene. Removal of bad motifs helps avoid problems such as poor PCR performance, poor expression levels, and low cloning efficiency. GENEius improves the properties of your genes to enhance expression and usability, while improving the probability of successfully assembling Genes or GeneStrands. Furthermore, flaws are removed and good motifs like new restriction sites are introduced in the gene. A list of good and bad motifs is available below:
GENEius introduces good motifs
- Distributes G's and C's across the whole sequence to increase synthetic feasibility
- Introduces useful restriction sites that can be used for subsequent sub-cloning projects
- Inserts motifs that can be useful for downstream applications (e.g., His6•Tag)
GENEius removes bad motifs
- Removes unwanted restriction sites
- Removes artificial splice sites
- Removes potential transcription factor binding sites
- Removes premature polyadenylation signals
- Removes inverted repeats or hairpins
Besides, optimizing the sequence to remove complexities, GENEius also helps in the design of Oligos that are used in the contruction of the Genes and GeneStrands. GENEius breaks the optimized (or unoptimized) sequence in silico into smaller oligos. The lengths and sequences of these individual oligos are optimized to provide higher success rates in the assembly of the Genes and GeneStrands, thereby, increasing the likelihood of sucess and a faster turnaround time.
Optimizing and ordering genes a decade ago was labor intensive and needed consultation. Today, you can simply use an online portal to enter your preferences and receive the optimal sequence.
GENEius is the online solution for fast and efficient ordering of synthetic genes. Conveniently enter your gene sequence in our Ecommerce system (Ecom) and GENEius will calculate the best possible sequence for your final expression vector. The integration of GENEius in the online shop creates a feature which allows the direct comparison of original and optimized sequence during the ordering process.
By integrating GENEius with our Order Wizard used for ordering Genes and GeneStrands, you receive several advantages:
- Sophisticated codon usage adaptation and sequence optimization
- Secure data transfer
- Fast and easy ordering
- Convenient quote request or submission of quotes
- Tracking of orders
- Order or quote history
- Individual project design
How to use GENEius for your project
GENEius is linked to our Ecom ordering system. When you choose codon usage adaption and optimisation of your gene sequences you can select the input codon usage table of your expression host and choose “bad motifs” like your cloning sites that will be excluded during adaption. You can even create your own bad motifs, e.g. transcription binding sites, artificial splice sites or polyadenylation signals. These sequence motifs will not be present in the optimised DNA sequence and therefore will not interfere with your downstream experiments.
Nature Is Biased! Each passing day gives us more insights into life at the molecular levels. Yet, several questions remain unanswered. What we know for certain today is that there is a codon bias; organims exhibit preferences for specific codons over other codons for encoding the same amino acid. This codon bias often leads to low heterologous expression, when the sequence is not optimized. In a human gene the amino acid arginine is mainly encoded by cgc and cgt.
| Arginine |
H. sapiens |
E. coli |
| CGT |
0.38 |
0.08 |
| CGC |
0.40 |
0.18 |
| CGA |
0.06 |
0.11 |
| CGG |
0.10 |
0.20 |
| AGA |
0.04 |
0.21 |
| AGG |
0.02 |
0.21 |
If E.coli is used as a host organism for gene expression, the unmodified human sequence will only yield minor amounts of proteins. This small yield reflects the limited number of tRNA available to translate the codons into the respective amino acid sequence. A vast amount of tRNAs are available if you optimise the codon usage—or better adapt the sequence of the template of origin to the host organism. Using the example above, selecting cgg, aga, and agg codons over cgt will ensure that there are sufficient amounts of tRNAs to produce high protein yields. Small effects make a big difference. The right balance in the codon usage of the host and the original organism's genome is critical for successful gene expression.We evaluate the performance of GENEius by adapting the sequence of a human gene for expression in Sf9 cells. The table below summarizes the features of the test gene before and after optimization.
| |
Best Conditions for expression in Sf9 |
Before GENEius optimization |
After GENEius optimization |
| GC content |
50% |
36.66% |
45.48% |
| Direct Repeats |
None |
9 |
None |
| Bad Motifs (e.g., PolyA or artificial splice sites) |
None |
1 PolyA signal present |
None |
| Bad Motifs (Cloning sites) |
Irrelevant |
Internal BamH1 site |
None |
| Good Motifs |
Irrelevant |
|
EcoRI at 5′ end + BamHI at 3′ end |
The composition of the sequence based on the optimal frequency for expression in Sf9 cells, the codons in the input sequence, and the codons frequency in the sequence after optimization by GENEius reveals the successful optimization of the input human gene sequence for expression in Sf9 cells.
Achieved Codon Usage
(Format: Optimal Frequency ∣ pre-optimization frequency ∣ Optimized Frequency) |
|---|
| D(10) |
E(11) |
F(9) |
G(24) |
| GAT: |
0.40 |
0.50 |
0.40 |
GAA: |
0.45 |
0.45 |
0.45 |
TTT: |
0.27 |
0.44 |
0.22 |
GGT: |
0.34 |
0.17 |
0.33 |
| GAC: |
0.60 |
0.50 |
0.60 |
GAG: |
0.55 |
0.55 |
0.55 |
TTC: |
0.73 |
0.56 |
0.78 |
GGC: |
0.31 |
0.33 |
0.29 |
| |
|
|
|
|
|
|
|
|
|
|
|
GGA: |
0.28 |
0.25 |
0.29 |
| |
|
|
|
|
|
|
|
|
|
|
|
GGG: |
0.07 |
0.25 |
0.08 |
| A(20) |
C(7) |
*(1) |
L(24) |
| GCT: |
0.35 |
0.25 |
0.35 |
TGT: |
0.39 |
0.43 |
0.43 |
TAA: |
0.63 |
0.00 |
1.00 |
TTA: |
0.10 |
0.08 |
0.08 |
| GCC: |
0.29 |
0.40 |
0.30 |
TGC: |
0.61 |
0.57 |
0.57 |
TAG: |
0.18 |
0.00 |
0.00 |
TTG: |
0.20 |
0.12 |
0.21 |
| GCA: |
0.18 |
0.25 |
0.20 |
|
|
|
|
TGA: |
0.18 |
1.00 |
0.00 |
CTT: |
0.12 |
0.12 |
0.12 |
| GCG: |
0.18 |
0.10 |
0.15 |
|
|
|
|
|
|
|
|
CTC: |
0.21 |
0.21 |
0.21 |
| |
|
|
|
|
|
|
|
|
|
|
|
CTA: |
0.09 |
0.08 |
0.08 |
| |
|
|
|
|
|
|
|
|
|
|
|
CTG: |
0.30 |
0.38 |
0.29 |
| M(11) |
N(16) |
H(4) |
I(12) |
| ATG: |
1.00 |
1.000 |
1.00 |
AAT: |
0.32 |
0.50 |
0.31 |
CAT: |
0.36 |
0.50 |
0.25 |
ATT: |
0.30 |
0.33 |
0.33 |
| |
|
|
|
AAC: |
0.68 |
0.50 |
0.69 |
CAC: |
0.64 |
0.50 |
0.75 |
ATC: |
0.54 |
0.50 |
0.50 |
| |
|
|
|
|
|
|
|
|
|
|
|
ATA: |
0.16 |
0.17 |
0.17 |
| K(23) |
T(18) |
W(7) |
V(25) |
| AAA: |
0.35 |
0.43 |
0.35 |
ACT: |
0.27 |
0.28 |
0.28 |
TGG: |
1.00 |
1.00 |
1.00 |
GTT: |
0.20 |
0.16 |
0.20 |
| AAG: |
0.65 |
0.57 |
0.65 |
ACC: |
0.32 |
0.33 |
0.33 |
|
|
|
|
GTC: |
0.29 |
0.24 |
0.28 |
| |
|
|
|
ACA: |
0.23 |
0.28 |
0.22 |
|
|
|
|
GTA: |
0.17 |
0.12 |
0.16 |
| |
|
|
|
ACG: |
0.18 |
0.11 |
0.17 |
|
|
|
|
GTG: |
0.34 |
0.48 |
0.36 |
| Q(6) |
P(10) |
S(15) |
R(22) |
| CAA: |
0.43 |
0.33 |
0.50 |
CCT: |
0.29 |
0.30 |
0.55 |
TCT: |
0.17 |
0.20 |
0.20 |
CGT: |
0.24 |
0.09 |
0.23 |
| CAG: |
0.57 |
0.67 |
0.50 |
CCC: |
0.28 |
0.30 |
0.55 |
TCC: |
0.21 |
0.20 |
0.20 |
CGC: |
0.30 |
0.18 |
0.32 |
| |
|
|
|
CCA: |
0.28 |
0.30 |
0.55 |
TCA: |
0.17 |
0.13 |
0.13 |
CGA: |
0.08 |
0.09 |
0.09 |
| |
|
|
|
CCG: |
0.16 |
0.10 |
0.55 |
TCG: |
0.13 |
0.07 |
0.13 |
CGG: |
0.00 |
0.18 |
0.00 |
| |
|
|
|
|
|
|
|
AGT: |
0.14 |
0.13 |
0.13 |
AGA: |
0.19 |
0.23 |
0.18 |
| |
|
|
|
|
|
|
|
AGC: |
0.18 |
0.27 |
0.20 |
AGG: |
0.19 |
0.23 |
0.18 |
| Y(6) |
| TAT: |
0.29 |
0.50 |
0.33 |
| TAC: |
0.71 |
0.50 |
0.67 |
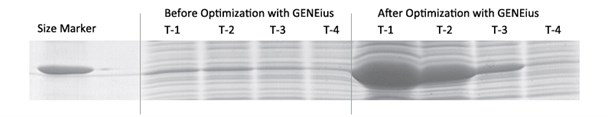
The gel below clearly demostrates that the expression level of the protein is much higher after sequence optimization by GENEius.