Site-Directed Mutageneses (SDM) are a collection of techniques that are used to mutate (add, change, or delete) specific nucleotides of a sequence in a plasmid. These techniques have allowed researchers to measure the relative importance of an amino acid at a specific position for the enzyme activity. By introducing mutations in the sequence, a researcher can change the nature of the amino acid in the protein; subsequent activity studies and comparison of with the activity of the wild-type (unmodified) enzyme allows for insights into the role of that amino acid in the enzyme activity. Besides being used to generate protein variants, SDM is used in several other contexts:
- Conventional Cloning – introduce or remove restriction sites
- Regulatory control – introduce or remove promoter/enhancer
- SNP (Single Nucleotide Polymorphism) Analysis – introduce SNPs in a plasmid
- CRISPR applications – removal of PAM sequence (where required)
The most widely used SDM technique for introducing a point mutation in a plasmid is PCR-based.
SDM using PCR
Typical steps to introduce mutations in a plasmid include:
- Design of a pair of mutagenesis primers
- PCR amplification of the template DNA (parent plasmid) using the designed primers
- Selectively digesting the parent plasmid using a methylation-dependent endonuclease
- Transformation of bacteria with the nuclease-resistant nicked plasmid
- Isolation of plasmids from the colonies
- Sequencing of plasmids obtained from individual colonies
- Sequence analysis to confirm the presence of desired mutation and absence of undesired/superfluous mutations.
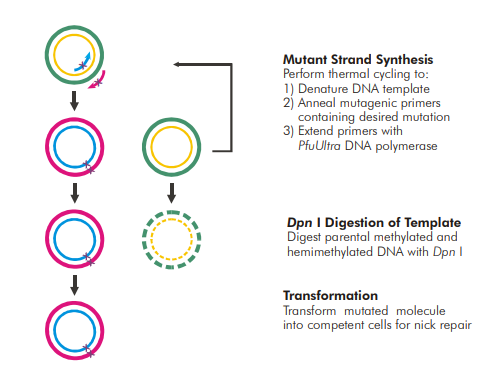
Image copied from Stratagene – QuickChange II
Design of PCR primers: Two mutagenesis primers are required for each SDM experiment – forward and reverse. A typical primer used in SDM experiments is between 25 and 45 bases long with a melting temperature Tm ³ 78 °C . To ensure that the primers anneal successfully to the plasmid, about 10 to 15 nt of complimentary sequence on either side of the desired mutation (typically 1–3 nt) is included in the mutagenesis primers. The presence of secondary structure, palindromic sequences, or other complexities in the mutagenesis primers must be evaluated and addressed. For more efficiency, the GC content must be over 40% and the 3¢-terminus must have one or more of C or G bases. The forward and reverse primers are usually complementary to each other. While overhangs in the primer pair are tolerated in these experiments, there must be a minimum of 6 bp overlap. Using these primers, mutations can only be introduced in the regions of sequence complimentary to the primers. The mutation carrying primers are incorporated into the PCR product, which eventually replaces the parent plasmid.