Highest specificity with Molecular Beacons
Molecular Beacons are hairpin shaped hybridisation probes with a 3‘ fluorescent dye and 5‘ quencher. This sequence structure makes Molecular Beacons highly specific and is therefore often used in clinical or diagnostic assays.
Product specifications:
- Available synthesis scales: 50 nmole - 1.0 µmole
- Probe lengths: 5-40 bases (wobbles non-defined ratio possible)
- HPLC purified by default
- TAT: 3–5 working days (1.0 µmole scale takes longer)
- Delivery format: dried or liquid at selected concentration
- QC report incl. ESI
A ready-to-use qPCR probe dilution buffer (10 mM Tris-HCl; 1 mM EDTA; pH 8) is provided along with all probes (1 ml for up to 3 probes).
Ordering
Molecular Beacons are ordered from the custom DNA order page by selecting the desired 5' and 3' modifications (dye-quencher combo). Go to custom order page.
Available dye-quencher combinations:
| 5' Reporter |
Abs [nm] |
Em [nm] |
3' Quencher |
FAM [FAM]
|
495 |
520 |
TAM, BHQ1, DAB, Eclip |
| TET [5TET] |
521 |
536 |
DAB |
| JOE [JOE] |
520 |
548 |
TAM |
| HEX [5HEX] |
535 |
556 |
BHQ1, DAB |
| Cyanine3 [Cy3] |
552 |
570 |
BHQ2, DAB |
| ROX [ROX] |
575 |
602 |
BHQ2 |
| Texas Red [TxRed] |
583 |
603 |
BHQ2 |
Cyanine5 [Cy5]
|
649 |
670 |
BHQ2, BBQ650 |
Principle of molecular beacons
Molecular beacons contain a fluorescent reporter and a quencher at their 5' and 3' ends, respectively.
The sequence of these probes is designed to allow the formation of a hairpin structure in which the fluorescent dye and the quencher are in close proximity.
A molecular beacon specific for the sequence of interest is used in PCR. The probe is designed to anneal between the PCR primers.
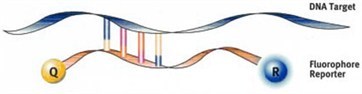
When the probe hybridises to its target sequence in the PCR annealing step, the loop opens and the fluorescent reporter and quencher are separated, resulting in a fluorescent signal upon excitation. The amount of signal is proportional to the amount of target sequence, and is measured in real time to allow quantification of the amount of target sequence.
Molecular beacon probe design
Molecular beacon probes are qPCR probes that are designed to create a stem-loop (hairpin) structure and contain a 5' dye and a 3' quencher. The loop is flanked by complimentary arms (usually 5-7 bp length) that form the stem when the arms hybridize with one other. Molecular beacon probes are extremely specific because precisely matched hybridization between the probe and target sequences is thermodynamically favored over the hairpin structure, and the hairpin structure is thermodynamically favored over hybridization to non-specific sequences [1,2]. The fact that hybridized molecular beacon probes are moved rather than degraded separates them from other forms of FRET-based qPCR probes. The processes below are performed for each PCR cycle, resulting in the identification of particular products.
- 1. The polymerase initiates DNA synthesis by extending from the primers.
- 2. During the annealing stage, the fluorescent dye (D) and quencher (Q) are separated by hybridization of molecular beacon probes to target sequences, resulting in fluorescence measured by the real-time PCR apparatus. Probes that do not hybridize to target DNA sequences reconstruct the hairpin structure and so do not glow.
- 3. The probe is shifted without being destroyed when the polymerase reaches the molecular beacon. As a result, molecular beacons can take part in numerous rounds of annealing.
- 4. The polymerase extends the primers farther to finish the synthesis of the DNA strand.
Additional Probe Options